class: center, middle # Many Types of Predictors  --- # We've Now Done Multiple Continuous Predictors `$$\large y_{i} = \beta_{0} + \sum \beta_{j}x_{ij} + \epsilon_{i}$$` `$$\large \epsilon_{i} \sim \mathcal{N}(0, \sigma)$$` --- # We've Previously Done One Categorical Variable with Many Levels `$$\large y_{ij} = \beta_{0} + \sum \beta_{j}x_{ij} + \epsilon_{ij}$$` `$$\large \epsilon_{ij} \sim \mathcal{N}(0, \sigma), \qquad x_{i} = 0,1$$` -- <br><br> (hey, wait, isn't that kinda the same model.... but where you can only belong to one level of one category?) --- # Now... Two Categories, Each with Many Levels, as Predictors `$$\large y_{ijk} = \beta_{0} + \sum \beta_{i}x_{ik} + \sum \beta_{j}x_{jk} + \epsilon_{ijk}$$` `$$\large \epsilon_{ijk} \sim N(0, \sigma^{2} ), \qquad x_{\_k} = 0,1$$` -- - This model is similar to MLR, but, now we multiple categories instead of multiple continuous predictors -- - This can be extended to as many categories as we want with linear algebra `$$Y = \beta X + \epsilon$$` --- class: center, middle  --- # Multiple Predictors: A Graphical View .center[ 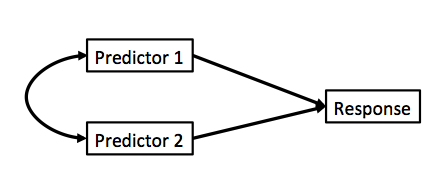 ] - Curved double-headed arrow indicates COVARIANCE between predictors that we account for. - We estimate the effect of each predictor **controlling** for all others. - Can be continous or categorical predictors --- # We Commonly Encounter Multiple Predictors in Randomized Controlled Blocked Designs .center[  ] --- # An Experiment with Orthogonal Treatments: A Graphical View .center[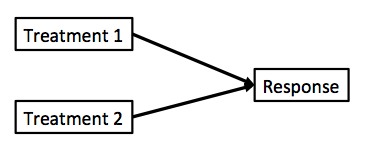] - Orthogonality breaks correlation - This is convenient for estimation - Observational data is not always so nice, which is OK! --- # Effects of Stickleback Density on Zooplankton <br><br> .pull-left[] .pull-right[] .bottom[Units placed across a lake so that 1 set of each treatment was ’blocked’ together] --- # Treatment and Block Effects <img src="many_types_of_predictors_files/figure-html/zooplankton_boxplot-1.png" style="display: block; margin: auto;" /> --- # Multiway Categorical Model - Many different treatment types - 2-Way is for Treatment and block - 3-Way for, e.g., Sticklebacks, Nutrients, and block - 4-way, etc., all possible -- - For experiments, we assume treatments are fully orthogonal - Each type of treatment type A has all levels of treatment type B - E.g., Each stickleback treatment is present in each block -- - Experiment is **balanced** for **simple effects** - Simple effect is the unique combination of two or more treatments - Balance implies the sample size for each treatment combination is the same - But, hey, this is more for inference, rather than **estimation** --- # Fitting a Model with Mutiple Categorical Predictors ``` r zoop_lm <- lm(zooplankton ~ treatment + block, data=zoop) zoop_lm ``` ``` Call: lm(formula = zooplankton ~ treatment + block, data = zoop) Coefficients: (Intercept) treatmenthigh treatmentlow block2 block3 3.420e+00 -1.640e+00 -1.020e+00 8.835e-16 -7.000e-01 block4 block5 -1.000e+00 -3.000e-01 ``` -- Note the treatment contrasts! --- # Assumptions of Categorical Models with Many Categories - Independence of data points - No relationship between fitted and residual values - **Additivity (linearity) of Treatments** - Normality within groups (of residuals) - Homoscedasticity (homogeneity of variance) of groups - No *collinearity* between levels of different categories --- # The Usual on Predictions <img src="many_types_of_predictors_files/figure-html/unnamed-chunk-2-1.png" style="display: block; margin: auto;" /> --- # Linearity (and additivity!) <img src="many_types_of_predictors_files/figure-html/unnamed-chunk-3-1.png" style="display: block; margin: auto;" /> --- # What is Non-Additivity? The effect of category depends on another - e.g. this grazing experiment <img src="many_types_of_predictors_files/figure-html/unnamed-chunk-4-1.png" style="display: block; margin: auto;" /> --- # Non-Additivity is Parabolic <img src="many_types_of_predictors_files/figure-html/unnamed-chunk-5-1.png" style="display: block; margin: auto;" /> --- class: center, middle  --- # Normality! <img src="many_types_of_predictors_files/figure-html/unnamed-chunk-6-1.png" style="display: block; margin: auto;" /> --- # HOV! <img src="many_types_of_predictors_files/figure-html/unnamed-chunk-7-1.png" style="display: block; margin: auto;" /> --- # Collinearity! <img src="many_types_of_predictors_files/figure-html/unnamed-chunk-8-1.png" style="display: block; margin: auto;" /> - by definition, not a problem in an experiment --- # How do We Understand the Modeled Results? - Coefficients (but treatment contrasts) - Expected means of levels of each category - Average over other categories - Differences between levels of each category --- # Coefficients and Treatment Contrasts <table class="table" style="color: black; margin-left: auto; margin-right: auto;"> <thead> <tr> <th style="text-align:left;"> term </th> <th style="text-align:right;"> estimate </th> <th style="text-align:right;"> std.error </th> </tr> </thead> <tbody> <tr> <td style="text-align:left;"> (Intercept) </td> <td style="text-align:right;"> 3.42 </td> <td style="text-align:right;"> 0.31 </td> </tr> <tr> <td style="text-align:left;"> treatmenthigh </td> <td style="text-align:right;"> -1.64 </td> <td style="text-align:right;"> 0.29 </td> </tr> <tr> <td style="text-align:left;"> treatmentlow </td> <td style="text-align:right;"> -1.02 </td> <td style="text-align:right;"> 0.29 </td> </tr> <tr> <td style="text-align:left;"> block2 </td> <td style="text-align:right;"> 0.00 </td> <td style="text-align:right;"> 0.37 </td> </tr> <tr> <td style="text-align:left;"> block3 </td> <td style="text-align:right;"> -0.70 </td> <td style="text-align:right;"> 0.37 </td> </tr> <tr> <td style="text-align:left;"> block4 </td> <td style="text-align:right;"> -1.00 </td> <td style="text-align:right;"> 0.37 </td> </tr> <tr> <td style="text-align:left;"> block5 </td> <td style="text-align:right;"> -0.30 </td> <td style="text-align:right;"> 0.37 </td> </tr> </tbody> </table> - Intercept is block 1, treatment control - Other coefs are all deviation from control in block 1 --- # Means Averaging Over Other Category ``` treatment emmean SE df lower.CL upper.CL control 3.02 0.205 8 2.548 3.49 high 1.38 0.205 8 0.908 1.85 low 2.00 0.205 8 1.528 2.47 Results are averaged over the levels of: block Confidence level used: 0.95 ``` ``` block emmean SE df lower.CL upper.CL 1 2.53 0.264 8 1.924 3.14 2 2.53 0.264 8 1.924 3.14 3 1.83 0.264 8 1.224 2.44 4 1.53 0.264 8 0.924 2.14 5 2.23 0.264 8 1.624 2.84 Results are averaged over the levels of: treatment Confidence level used: 0.95 ``` --- # Visualize the Expected Means <img src="many_types_of_predictors_files/figure-html/unnamed-chunk-10-1.png" style="display: block; margin: auto;" /> --- # Differences Between Treatments ``` contrast estimate SE df lower.CL upper.CL control - high 1.64 0.289 8 0.972 2.3075 control - low 1.02 0.289 8 0.352 1.6875 high - low -0.62 0.289 8 -1.288 0.0475 Results are averaged over the levels of: block Confidence level used: 0.95 ``` --- # Many Additive Predictors 1. Multiple Linear Regression 2. Many Categories with Many Levels 3. <font color = "red">Combining Categorical and Continuous Predictors</font> --- # It's All One **The Linear Model** `$$\boldsymbol{Y} = \boldsymbol{b X} + \boldsymbol{\epsilon}$$` -- **Multiple Continuous Predictors** `$$y_{i} = \beta_{0} + \sum \beta_{j}x_{ij} + \epsilon_{i}$$` -- **Many Categorical Predictors** `$$y_{ijk} = \beta_{0} + \sum \beta_{i}x_{ik} + \sum \beta_{j}x_{jk} + \epsilon_{ijk}$$` --- # Mixing Continuous and Categorical Predictors: Analysis of Covariance `$$y_{ij} = \beta_0 + \beta_{1}x_{1i} + \sum\beta_j x_{ij} + + \epsilon_{ij}$$` $$ x_{ij} = 0,1 \qquad \epsilon \sim \mathcal{N}(0,\sigma)$$ - Categorical Variable + a continuous predictor - Often used to correct for a gradient or some continuous variable affecting outcome - OR used to correct a regression due to additional groups that may throw off slope estimates - e.g. Simpson's Paradox: A positive relationship between test scores and academic performance can be masked by gender differences --- # Simpson's Paradox 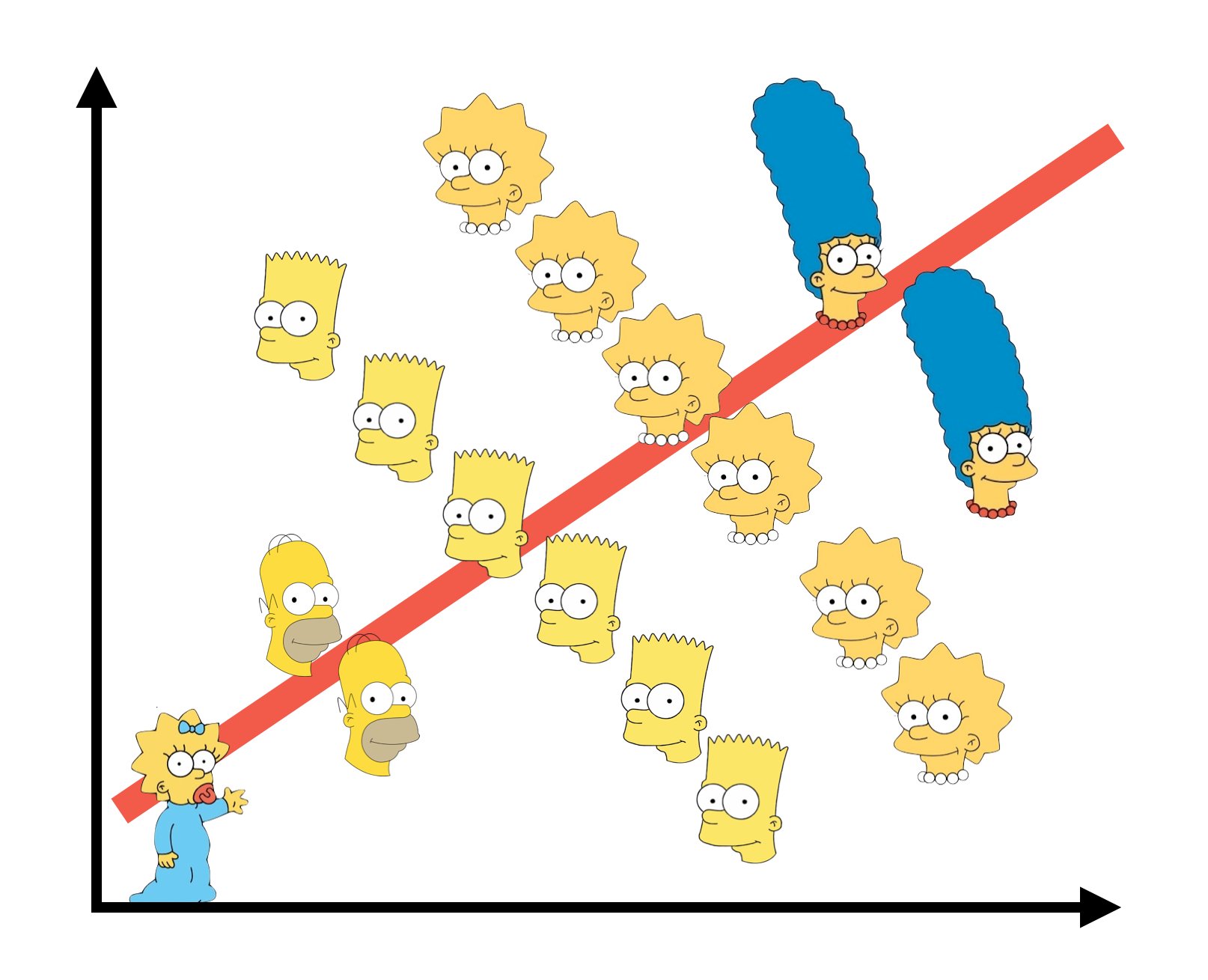 --- # What is Simpson's Paradox: Penguin Example <img src="many_types_of_predictors_files/figure-html/unnamed-chunk-12-1.png" style="display: block; margin: auto;" /> --- # What is Simpson's Paradox: Penguin Example <img src="many_types_of_predictors_files/figure-html/unnamed-chunk-13-1.png" style="display: block; margin: auto;" /> -- Note: This can happen with just continuous variables as well --- # Neanderthals and Categorical/Continuous Variables .center[  ] Who had a bigger brain: Neanderthals or us? --- # The Means Look the Same... <img src="many_types_of_predictors_files/figure-html/neand_boxplot-1.png" style="display: block; margin: auto;" /> --- # But there appears to be a Relationship Between Body and Brain Mass <img src="many_types_of_predictors_files/figure-html/neand_plot-1.png" style="display: block; margin: auto;" /> --- # And Mean Body Mass is Different <img src="many_types_of_predictors_files/figure-html/neand_boxplot2-1.png" style="display: block; margin: auto;" /> --- class: center, middle  --- # Categorical Model with a Continuous Covariate for Control <img src="many_types_of_predictors_files/figure-html/neand_plot_fit-1.png" style="display: block; margin: auto;" /> Evaluate a categorical effect(s), controlling for a *covariate* (parallel lines) Groups modify the *intercept*. --- # Assumptions are the Same! - Independence of data points - Additivity of Treatment and Covariate (Parallel Slopes) - Normality and homoscedacticity within groups (of residuals) - No relationship between fitted and residual values --- # Linearity Assumption KEY <img src="many_types_of_predictors_files/figure-html/zoop_assumptions-1.png" style="display: block; margin: auto;" /> --- # Test for Parallel Slopes We fit a model where slopes are not parallel: `$$y_{ijk} = \beta_0 + \beta_{1}x_1 + \sum_{j}^{i=1}\beta_j x_{ij} + \sum_{j}^{i=1}\beta_{k}x_1 x_{ij} + \epsilon_ijk$$` -- <table class="table" style="color: black; margin-left: auto; margin-right: auto;"> <thead> <tr> <th style="text-align:left;"> term </th> <th style="text-align:right;"> estimate </th> <th style="text-align:right;"> std.error </th> </tr> </thead> <tbody> <tr> <td style="text-align:left;"> (Intercept) </td> <td style="text-align:right;"> 4.2534953 </td> <td style="text-align:right;"> 0.9768911 </td> </tr> <tr> <td style="text-align:left;"> speciesrecent </td> <td style="text-align:right;"> 1.1809274 </td> <td style="text-align:right;"> 1.0623095 </td> </tr> <tr> <td style="text-align:left;"> lnmass </td> <td style="text-align:right;"> 0.7135471 </td> <td style="text-align:right;"> 0.2270081 </td> </tr> <tr> <td style="text-align:left;"> speciesrecent:lnmass </td> <td style="text-align:right;"> -0.2594885 </td> <td style="text-align:right;"> 0.2481073 </td> </tr> </tbody> </table> -- If you have an interaction, welp, that's a different model - slopes vary by group! --- # VIF Also *Very* Important <img src="many_types_of_predictors_files/figure-html/unnamed-chunk-14-1.png" style="display: block; margin: auto;" /> --- # Usual Normality Assumption <img src="many_types_of_predictors_files/figure-html/unnamed-chunk-15-1.png" style="display: block; margin: auto;" /> --- # Usual HOV Assumption <img src="many_types_of_predictors_files/figure-html/unnamed-chunk-16-1.png" style="display: block; margin: auto;" /> --- # Usual Outlier Assumption <img src="many_types_of_predictors_files/figure-html/unnamed-chunk-17-1.png" style="display: block; margin: auto;" /> --- # The Results - We can look at coefficients - We can look at means adjusted for covariate - Visualize! Visualize! Visualize! --- # Those Coefs <table class="table" style="color: black; margin-left: auto; margin-right: auto;"> <thead> <tr> <th style="text-align:left;"> term </th> <th style="text-align:right;"> estimate </th> <th style="text-align:right;"> std.error </th> </tr> </thead> <tbody> <tr> <td style="text-align:left;"> (Intercept) </td> <td style="text-align:right;"> 5.19 </td> <td style="text-align:right;"> 0.40 </td> </tr> <tr> <td style="text-align:left;"> speciesrecent </td> <td style="text-align:right;"> 0.07 </td> <td style="text-align:right;"> 0.03 </td> </tr> <tr> <td style="text-align:left;"> lnmass </td> <td style="text-align:right;"> 0.50 </td> <td style="text-align:right;"> 0.09 </td> </tr> </tbody> </table> - Intercept is species = neanderthal, but lnmass = 0? - Categorical coefficient is change in intercept for recent - lnmass coefficient is change in ln brain mass per change in 1 unit of ln mass --- # Groups Means at Mean of Covariate <table class="table" style="color: black; margin-left: auto; margin-right: auto;"> <thead> <tr> <th style="text-align:left;"> species </th> <th style="text-align:right;"> emmean </th> <th style="text-align:right;"> SE </th> <th style="text-align:right;"> df </th> <th style="text-align:right;"> lower.CL </th> <th style="text-align:right;"> upper.CL </th> </tr> </thead> <tbody> <tr> <td style="text-align:left;"> neanderthal </td> <td style="text-align:right;"> 7.272 </td> <td style="text-align:right;"> 0.024 </td> <td style="text-align:right;"> 36 </td> <td style="text-align:right;"> 7.223 </td> <td style="text-align:right;"> 7.321 </td> </tr> <tr> <td style="text-align:left;"> recent </td> <td style="text-align:right;"> 7.342 </td> <td style="text-align:right;"> 0.013 </td> <td style="text-align:right;"> 36 </td> <td style="text-align:right;"> 7.317 </td> <td style="text-align:right;"> 7.367 </td> </tr> </tbody> </table> Can also evaluate for other levels of the covariate as is interesting --- # We Can Compare Groups Adjusting for Covariates .center[  ] --- # Difference Between Groups at Mean of Covariate <table class="table" style="color: black; margin-left: auto; margin-right: auto;"> <thead> <tr> <th style="text-align:left;"> contrast </th> <th style="text-align:right;"> estimate </th> <th style="text-align:right;"> SE </th> <th style="text-align:right;"> df </th> <th style="text-align:right;"> lower.CL </th> <th style="text-align:right;"> upper.CL </th> </tr> </thead> <tbody> <tr> <td style="text-align:left;"> neanderthal - recent </td> <td style="text-align:right;"> -0.07 </td> <td style="text-align:right;"> 0.028 </td> <td style="text-align:right;"> 36 </td> <td style="text-align:right;"> -0.128 </td> <td style="text-align:right;"> -0.013 </td> </tr> </tbody> </table> --- # Visualizing Result Says it All! <img src="many_types_of_predictors_files/figure-html/unnamed-chunk-19-1.png" style="display: block; margin: auto;" /> --- # Or Plot at the Mean Level of the Covariate <img src="many_types_of_predictors_files/figure-html/unnamed-chunk-20-1.png" style="display: block; margin: auto;" /> --- # Extensions of the Linear Additive Model - Wow, we sure can fit a lot in there! - Categorical is just continuous as 0/1 - So, we can build a LOT of models, limited only by our imagination! -- - But what was that nonadditive thing again...? Wait a minuite...