class: center, middle # Generalized Linear Models  <style type="text/css"> .pull-left-small { float: left; width: 20%; } .pull-right-large { float: right; width: 80%; } </style> --- background-image:url("images/glm/4_consumers.png") background-size: contain --- background-image:url("images/glm/plate.png") background-size: contain --- ### My Data <table class=" lightable-paper" style='font-family: "Arial Narrow", arial, helvetica, sans-serif; margin-left: auto; margin-right: auto;'> <thead> <tr> <th style="text-align:right;"> predator_richness </th> <th style="text-align:right;"> percent_open </th> </tr> </thead> <tbody> <tr> <td style="text-align:right;"> 1 </td> <td style="text-align:right;"> 81 </td> </tr> <tr> <td style="text-align:right;"> 2 </td> <td style="text-align:right;"> 100 </td> </tr> <tr> <td style="text-align:right;"> 2 </td> <td style="text-align:right;"> 70 </td> </tr> <tr> <td style="text-align:right;"> 1 </td> <td style="text-align:right;"> 97 </td> </tr> <tr> <td style="text-align:right;"> 2 </td> <td style="text-align:right;"> 58 </td> </tr> <tr> <td style="text-align:right;"> 1 </td> <td style="text-align:right;"> 96 </td> </tr> <tr> <td style="text-align:right;"> 4 </td> <td style="text-align:right;"> 92 </td> </tr> <tr> <td style="text-align:right;"> 1 </td> <td style="text-align:right;"> 83 </td> </tr> <tr> <td style="text-align:right;"> 2 </td> <td style="text-align:right;"> 99 </td> </tr> <tr> <td style="text-align:right;"> 2 </td> <td style="text-align:right;"> 92 </td> </tr> </tbody> </table> --- ## So.... <img src="generalized_linear_models_files/figure-html/cons_plot-1.png" style="display: block; margin: auto;" /> --- ## That was a Terrible Idea <img src="generalized_linear_models_files/figure-html/unnamed-chunk-2-1.png" style="display: block; margin: auto;" /> --- ## That was a Terrible Idea <img src="generalized_linear_models_files/figure-html/unnamed-chunk-3-1.png" style="display: block; margin: auto;" /> --- ## That was a Terrible Idea <img src="generalized_linear_models_files/figure-html/unnamed-chunk-4-1.png" style="display: block; margin: auto;" /> --- ## Log (and other transformations) are not better <img src="generalized_linear_models_files/figure-html/unnamed-chunk-5-1.png" style="display: block; margin: auto;" /> --- ## The Answer is Not Normal.... or Linear <img src="generalized_linear_models_files/figure-html/unnamed-chunk-6-1.png" style="display: block; margin: auto;" /> --- class:center, middle  --- # A Generalized Linear World 1. The Generalized Linear Model 2. Logistic Regression & Assumption Tests 3. Logit Curves with Continuous Data --- # We Have Lived In a Normal World - Until Now <img src="generalized_linear_models_files/figure-html/unnamed-chunk-7-1.png" style="display: block; margin: auto;" /> --- # Gaussian Distributions are Part of a Larger Interconnected Family of Distributions  --- # It Gets Crazy .center[  ] See https://www.randomservices.org/random/special/GeneralExponential.html --- # The Linear Model with Gaussian Error $$\Large \hat{Y_i} = \boldsymbol{\beta X_i} $$ <br><br> `$$\Large Y_i \sim \mathcal{N}(\hat{Y_i},\sigma^{2})$$` --- # Let's Make a Small Change $$\Large \boldsymbol{\eta_{i}} = \boldsymbol{\beta X_i} $$ <br><br> `$$\Large \hat{Y_i} = \eta_{i}$$` <br><br> `$$\Large Y_i \sim \mathcal{N}(\hat{Y_i},\sigma^{2})$$` --- # Another Teeeny Change - Link Functions $$\Large \boldsymbol{\eta_{i}} = \boldsymbol{\beta X_i} $$ <br><br> `$$\Large f(\hat{Y_i}) = \eta_{i}$$` .red[f is a link function] .red[for linear regression, we call this an **identity link**] `$$\Large Y_i \sim \mathcal{N}(\hat{Y_i},\sigma^{2})$$` --- ## Hold up, isn't this just transformation? - No. -- - `\(log(Y) = \beta X + \epsilon\)` means that `\(Y = exp(\beta X + \epsilon)\)` -- - A **log link** with a normal residual means that - `\(log(\widehat{Y}) = \beta X\)` but `\(\epsilon\)` is additive to `\(\widehat{Y}\)` - SO `\(Y = exp(\beta X) + \epsilon\)` --- # So if THIS is the Generalized Linear Model with a Gaussian Error and an Identity Link... $$\Large \boldsymbol{\eta_{i}} = \boldsymbol{\beta X_i} $$ <br><br> `$$\Large\hat{Y_i} = \eta_{i}$$` <br><br> `$$\Large Y_i \sim \mathcal{N}(\hat{Y_i},\sigma^{2})$$` --- # The Generalized Linear Model $$\Large \boldsymbol{\eta_{i}} = \boldsymbol{\beta X_i} $$ <br><br> `$$\Large f(\hat{Y_i}) = \eta_{i}$$` .red[our link doesn't have to be linear] <br><br> `$$\Large Y_i \sim \mathcal{D}(\hat{Y_i}, \theta)$$` <br><br> `\(\mathcal{D}\)` is some distribution from the **exponential family**, and `\(\theta\)` are additional parameters specifying dispersion --- # What Are Properties of Data that is Not Normal? -- - Bounded -- - Discrete -- - Variance scales with mean -- - Skewed, bimodal, or kurtotic --- class: center, middle  --- # Count Data - Poisson or Negative Binomial Discrete, variance-mean relationships (linear or squared) <img src="generalized_linear_models_files/figure-html/unnamed-chunk-9-1.png" style="display: block; margin: auto;" /> --- # Outcomes of Trials - Binomial Discrete, Yes/No or # of Outcomes of a Class - we model the probability .pull-left-small[  ] .pull-right-large[ <img src="generalized_linear_models_files/figure-html/unnamed-chunk-10-1.png" style="display: block; margin: auto;" /> ] --- # Time to Achieve Outcomes - Gamma Skewed continuous, variance-mean relationship .pull-left-small[  ] .pull-right-large[ <img src="generalized_linear_models_files/figure-html/unnamed-chunk-11-1.png" style="display: block; margin: auto;" /> ] --- # Percents and Porportions - Beta Bounded, Unimodal to Bimodal .pull-left-small[  ] .pull-right-large[ <img src="generalized_linear_models_files/figure-html/unnamed-chunk-12-1.png" style="display: block; margin: auto;" /> ] --- # The Generalized Linear Model $$\Large \boldsymbol{\eta_{i}} = \boldsymbol{\beta X_i} $$ <br><br> `$$\Large f(\hat{Y_i}) = \eta_{i}$$` .red[f is a link function] <br><br> `$$\Large Y_i \sim \mathcal{D}(\hat{Y_i}, \theta)$$` <br><br> `\(\mathcal{D}\)` is some distribution from the **exponential family**, and `\(\theta\)` are additional parameters specifying dispersion --- # Generalized Linear Models: Distributions 1. The error distribution is from the **exponential family** - e.g., Normal, Poisson, Binomial, and more. -- 2. For these distributions, the variance is a funciton of the fitted value on the curve: `\(var(Y_i) = \theta V(\hat{Y_i})\)` - For a normal distribution, `\(var(Y_i) = \theta*1\)` as `\(V(\hat{Y_i})=1\)` - For a poisson distribution, `\(var(Y_i) = 1*\mu_i\)` as `\(V(\hat{Y_i})=\hat{Y_i}\)` -- 3. They have *canonical links* which are link functions that fall out from the shape of the distribution and map on to the domain of possible values - e.g., the identity for the Gaussian - We can also use non-canonical links --- # Distributions, Canonical Links, and Dispersion |Distribution | Canonical Link | Variance Function| |-------------|-------------|-------------| |Normal | identity | `\(\theta\)`| |Poisson | log | `\(\hat{Y_i}\)`| |Binomial | logit | `\(\hat{Y_i}(1-\hat{Y_i})\)`| |Negative Binomial | log | `\(\hat{Y_i} + \theta\hat{Y_i}^2\)`| |Gamma | inverse | `\(\hat{Y_i}^2\)`| | Beta | logit | `\(\hat{Y_i}(1 - \hat{Y_i})/(\theta + 1)\)`| The key is to think about your residual variance and what is appropriate --- # A Generalized Linear World 1. The Generalized Linear Model 2. .red[Logistic Regression & Assumption Tests] 3. Logit Curves with Continuous Data --- background-image: url("images/25/cryptosporidiosis-cryptosporidium-hominis.jpeg") background-position: center background-size: cover class: inverse, center # Cryptosporidium Parasite --- background-image: url("images/25/mouseinject.jpg") background-position: center background-size: cover class: inverse # What dose is needed to generate infection? --- # Cryptosporidum Infection Rates <img src="generalized_linear_models_files/figure-html/crypto_plot-1.png" style="display: block; margin: auto;" /> --- # This is not linear or gaussian <img src="generalized_linear_models_files/figure-html/crypto_linear-1.png" style="display: block; margin: auto;" /> --- class: center, middle # STOP - before we go any further, you COULD .red[logit transform] a fraction from trials and use a linear model - if it meets assumptions, OK! --- # Logit Transform  --- # Why GLM then? 1. It might not work, 2. Unequal # of trials per point, 3. A binary response <img src="generalized_linear_models_files/figure-html/unnamed-chunk-13-1.png" style="display: block; margin: auto;" /> --- # Binary GLM <img src="generalized_linear_models_files/figure-html/unnamed-chunk-14-1.png" style="display: block; margin: auto;" /> --- class: center, middle  --- # Binomial Distribution (coin flips!) $$ Y_i \sim B(prob, size) $$ * Discrete Distribution -- * prob = probability of something happening (% Infected) -- * size = # of discrete trials -- * Used for frequency or probability data -- * We estimate coefficients that influence *prob* --- # Generalized Linear Model with a Logit Link $$\Large \boldsymbol{\eta_{i}} = \boldsymbol{\beta X_i} $$ <br><br><br> `$$\Large Logit(\hat{Y_i}) = \eta_{i}$$` .red[Logit Link Function] <br><br><br> `$$\Large Y_i \sim \mathcal{B}(\hat{Y_i}, size)$$` --- # Generalized Linear Model with Logit Link ```r crypto_glm <- glm(Y/N ~ Dose, weights=N, family=binomial(link="logit"), data=crypto) ``` OR, with Success and Failures ```r crypto_glm <- glm(cbind(Y, Y-N) ~ Dose, family=binomial(link="logit"), data=crypto) ``` --- # How About those Assumptions? 1. Is there a match between observations and predictions? 2. No Outliers 3. Quantile Residuals --- # How About those Assumptions? <img src="generalized_linear_models_files/figure-html/unnamed-chunk-15-1.png" style="display: block; margin: auto;" /> --- # How About those Assumptions? <img src="generalized_linear_models_files/figure-html/unnamed-chunk-16-1.png" style="display: block; margin: auto;" /> --- # Assessing Assumptions when Modeling Variance: Quantile Residuals - Every residual point comes from a distribution -- - Every distribution has a *cummulative distribution function* -- - The CDF of a probability distribution defines it's quantiles - from 0 to 1. -- - We can determine the quantile of any point relative to its distribution --- # A Quantile from a Distribution: Say this is your distribution <img src="generalized_linear_models_files/figure-html/unnamed-chunk-17-1.png" style="display: block; margin: auto;" /> --- # A Quantile from a Distribution: Overlay the CDF <img src="generalized_linear_models_files/figure-html/unnamed-chunk-18-1.png" style="display: block; margin: auto;" /> --- # A Quantile from a Distribution: This is our Point <img src="generalized_linear_models_files/figure-html/unnamed-chunk-19-1.png" style="display: block; margin: auto;" /> --- # A Quantile from a Distribution: We see where it lies on the CDF <img src="generalized_linear_models_files/figure-html/unnamed-chunk-20-1.png" style="display: block; margin: auto;" /> --- # A Quantile from a Distribution: And we can get its quantile <img src="generalized_linear_models_files/figure-html/unnamed-chunk-21-1.png" style="display: block; margin: auto;" /> --- # Randomized quantile residuals - Every observation is distributed around an estimated value -- - We can calculate its quantile -- - If model fits well, quantiles of residuals should be uniformly distributed -- - I.E., for any point, if we had its distribution, there should be no bias in its quantile -- - We do this via simulation for flexibility -- - Works for **many** (any?) models --- # Randomized quantile residuals: Steps 1. Get 1000 (or more!) simulations of model coefficients -- 2. For each response (y) value, create an empirical distribution from the simuations -- 3. For each response, determine it's quantile from that empirical distribution -- 4. The quantiles of all y values should be uniformly distributed - QQ plot of a uniform distribution! --- # Randomized quantile residuals: Visualize <img src="generalized_linear_models_files/figure-html/rqr-1.png" style="display: block; margin: auto;" /> --- # Randomized quantile residuals: Visualize <img src="generalized_linear_models_files/figure-html/rqr1-1.png" style="display: block; margin: auto;" /> --- # Randomized quantile residuals: Visualize <img src="generalized_linear_models_files/figure-html/rqr2-1.png" style="display: block; margin: auto;" /> --- # Randomized quantile residuals: Visualize <img src="generalized_linear_models_files/figure-html/rqr3-1.png" style="display: block; margin: auto;" /> --- # Randomized quantile residuals: Visualize <img src="generalized_linear_models_files/figure-html/rqr4-1.png" style="display: block; margin: auto;" /> --- # Randomized Quantile Residuals from Squid Model <img src="generalized_linear_models_files/figure-html/unnamed-chunk-22-1.png" style="display: block; margin: auto;" /> --- # OK, What Are We Estimating? ### The Odds ## The Odds `$$Odds = \frac{p}{1-p}$$` (e.g., 50:50 is an odds ratio of 1, 2:1 odds are pretty good! 1:100 odds is not!) -- Logit coefficients are in: `$$Log-Odds = Log\frac{p}{1-p} = logit(p)$$` (log of 1 is 0, log of .69, log of .01 is -4.60517) --- # Coefficients and Interpretation <table class="table" style="margin-left: auto; margin-right: auto;"> <thead> <tr> <th style="text-align:left;"> term </th> <th style="text-align:right;"> estimate </th> <th style="text-align:right;"> std.error </th> </tr> </thead> <tbody> <tr> <td style="text-align:left;"> (Intercept) </td> <td style="text-align:right;"> -1.408 </td> <td style="text-align:right;"> 0.148 </td> </tr> <tr> <td style="text-align:left;"> Dose </td> <td style="text-align:right;"> 0.013 </td> <td style="text-align:right;"> 0.001 </td> </tr> </tbody> </table> -- - Woof. Logit coefficients are tricky (go visualization!) -- - In essence, an increase in 1 unit of dose = an exp( 0.0135) increase in the ODDS of infection - 1.014 increase (remember, 1 is a 50:50 chance!) -- - You *can* use this to estimate change in %, but it requires knowing where you start from on the curve (this is a nonlinear curve after all) --- # Easier to visualize to interpret <img src="generalized_linear_models_files/figure-html/crypto_logit-1.png" style="display: block; margin: auto;" /> --- # Final Note on Logistic Regression - Ideal for yes/no classification - e.g. if your response variable is purely binary - Has an extension to *multinomial logistic regression* - Many categories, often used for classification - Only use if you can represent data as indepenent *trials* - Internally, breaks responses into yes/no --- # A Generalized Linear World 1. The Generalized Linear Model 2. Logistic Regression & Assumption Tests 3. .red[Logit Curves with Continuous Data] --- background-image:url("images/glm/plate.png") background-size: contain --- ### My Data <table class=" lightable-paper" style='font-family: "Arial Narrow", arial, helvetica, sans-serif; margin-left: auto; margin-right: auto;'> <thead> <tr> <th style="text-align:right;"> predator_richness </th> <th style="text-align:right;"> percent_open </th> </tr> </thead> <tbody> <tr> <td style="text-align:right;"> 1 </td> <td style="text-align:right;"> 81 </td> </tr> <tr> <td style="text-align:right;"> 2 </td> <td style="text-align:right;"> 100 </td> </tr> <tr> <td style="text-align:right;"> 2 </td> <td style="text-align:right;"> 70 </td> </tr> <tr> <td style="text-align:right;"> 1 </td> <td style="text-align:right;"> 97 </td> </tr> <tr> <td style="text-align:right;"> 2 </td> <td style="text-align:right;"> 58 </td> </tr> <tr> <td style="text-align:right;"> 1 </td> <td style="text-align:right;"> 96 </td> </tr> <tr> <td style="text-align:right;"> 4 </td> <td style="text-align:right;"> 92 </td> </tr> <tr> <td style="text-align:right;"> 1 </td> <td style="text-align:right;"> 83 </td> </tr> <tr> <td style="text-align:right;"> 2 </td> <td style="text-align:right;"> 99 </td> </tr> <tr> <td style="text-align:right;"> 2 </td> <td style="text-align:right;"> 92 </td> </tr> </tbody> </table> --- # This was wrong <img src="generalized_linear_models_files/figure-html/cons_plot-1.png" style="display: block; margin: auto;" /> --- # Dirchlet (and Beta) Regression  - Dirchlet distribution describes probability density for **many** variables that are constrained to sum together to 1. - Beta distribution is a special case of the Dirchlet where there are only two variables that must sum to one. For example: - Percent cover - Completion rates - Purity of distillate --- # Percents and Porportions - Beta Bounded, Unimodal to Bimodal .pull-left-small[  ] .pull-right-large[ <img src="generalized_linear_models_files/figure-html/unnamed-chunk-24-1.png" style="display: block; margin: auto;" /> ] --- # The Beta Generalized Linear Model $$\Large \boldsymbol{\eta_{i}} = \boldsymbol{\beta X_i} $$ <br><br> `$$\Large f(\hat{Y_i}) = \eta_{i}$$` .red[canonical link is logit, so] `\(0 < \hat{Y_i} < 1\)` <br><br> `$$\Large Y_i \sim \mathcal{B}(\hat{Y_i}, \phi)$$` <br><br> `\(\mathcal{D}\)` is the Beta Distribution and `\(\phi\)` is a parameter that changes with the mean so that `\(Var(Y) = \hat{Y_i}(1-\hat{Y_i})/(1+\phi)\)` --- # A Few Other Links 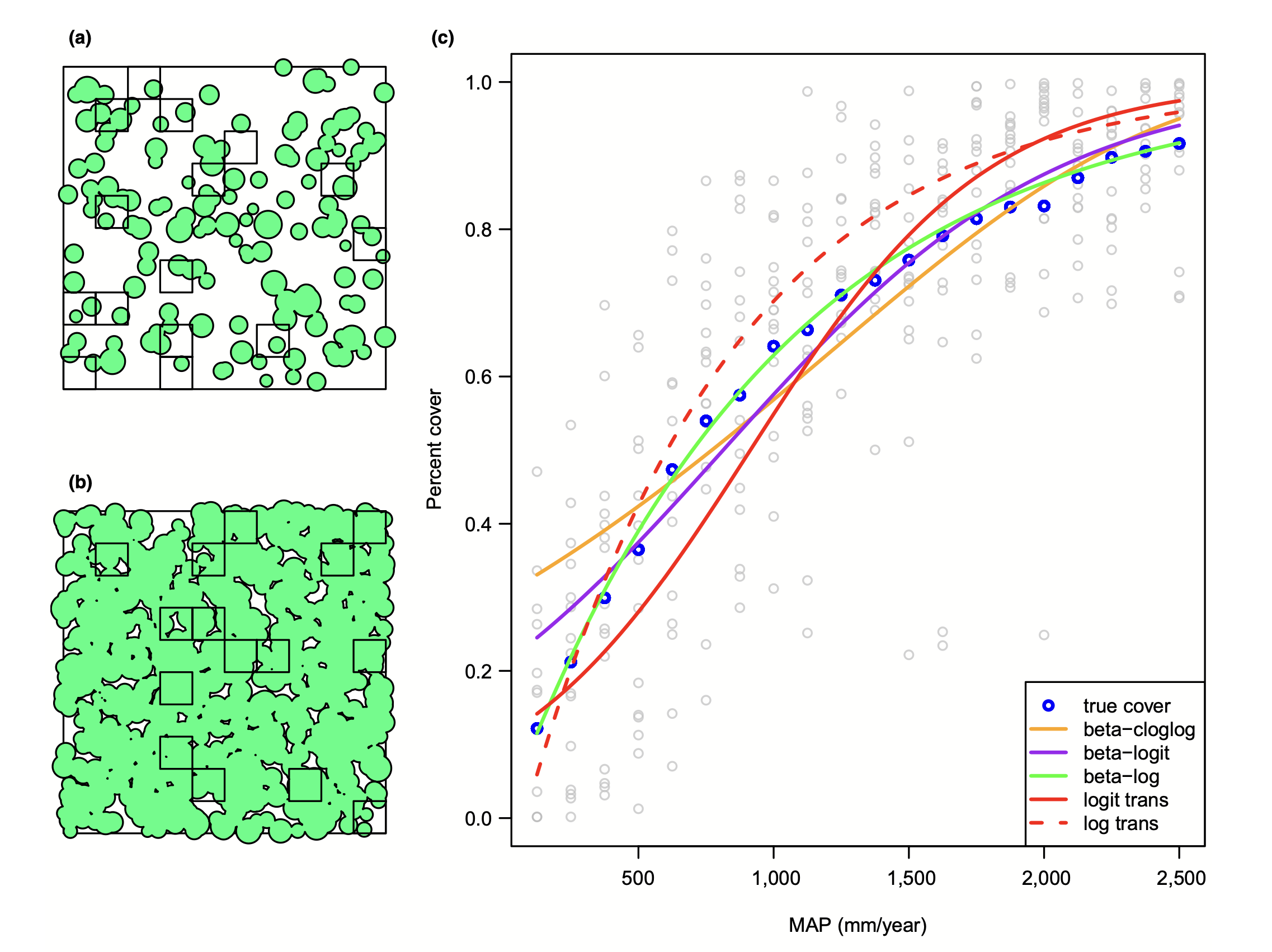 --- ## Our Model ```r library(glmmTMB) cons_div_aug <- cons_div_aug |> mutate(porportion_open = percent_open/100) breg <- glmmTMB(porportion_open ~ predator_richness, data = cons_div_aug, family = ordbeta()) ``` (note, ordbeta allows for 0, 1 - ordered beta regression) --- # Check Your Quantile Residuals! <img src="generalized_linear_models_files/figure-html/unnamed-chunk-26-1.png" style="display: block; margin: auto;" /> --- # Let's Plot Instead of Coefficients <img src="generalized_linear_models_files/figure-html/unnamed-chunk-27-1.png" style="display: block; margin: auto;" /> --- # The GLM Frontier .pull-left[  ] .pull-right[ - GLMs can be intimidating, but, think about your data and shape of your data generating process - They are a jumping off point to more interesting error distributions - Mastering GLMs will unlock data that formerly was just frustrating ]