Merging Data
- Problem: I have two data sets
- One is biological information
- One is physical information
- They have a common key - e.g., Lat/Long
Join
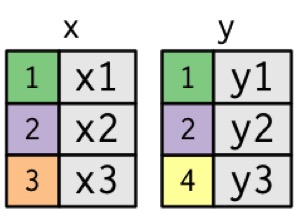
The data
tibble [98 × 11] (S3: tbl_df/tbl/data.frame)
$ Stand : chr [1:98] "Athol 1" "Athol 2" "Athol 4" "Athol 6" ...
$ Year : num [1:98] 2003 2003 2003 2003 2004 ...
$ Longitude : num [1:98] -72.2 -72.2 -72.2 -72.2 -72.1 ...
$ Latitude : num [1:98] 42.5 42.5 42.5 42.6 42.6 ...
$ Live BA : num [1:98] 36.3 31.2 35.9 32.6 23 ...
$ Dead Hem BA : num [1:98] 0.46 0.46 0 0 0 2.87 0 0 0 1.15 ...
$ Hem Vigor : num [1:98] 1.6 1.18 1.47 1.86 1.25 1.9 1.91 1.56 1 1.81 ...
$ Hem Den : num [1:98] 1450 1250 900 725 600 725 825 450 400 925 ...
$ Dead Hem Den : num [1:98] 50 50 0 50 0 150 50 50 0 100 ...
$ Tree Den : num [1:98] 2125 1725 1700 1100 1075 ...
$ Borer Density: num [1:98] 0 0 0 0 0 0 0 0 0 0 ...Environmental Information
tibble [111 × 11] (S3: tbl_df/tbl/data.frame)
$ Stand : chr [1:111] "Athol 1" "Athol 2" "Athol 3" "Athol 4" ...
$ Year : num [1:111] 2003 2003 2003 2003 2003 ...
$ Mapped Code: chr [1:111] "A" "A" "A" "B" ...
$ Aspect : num [1:111] 213.2 357 292.5 80.5 227.5 ...
$ Slope : num [1:111] 3.8 27.83 23.83 8.78 12.17 ...
$ Longitude : num [1:111] -72.2 -72.2 -72.2 -72.2 -72.2 ...
$ Latitude : num [1:111] 42.5 42.5 42.6 42.5 42.6 ...
$ Elevation : num [1:111] 269 220 231 247 233 ...
$ Area : num [1:111] 35.8 36.6 33.7 94.7 40.7 ...
$ Humus : num [1:111] 9.9 5.92 5.58 6.89 3.71 5.25 7.33 12.4 6.75 8.85 ...
$ Logged : num [1:111] 1 1 1 1 1 1 1 0 1 1 ...The problem
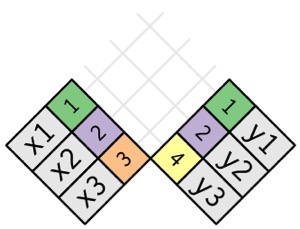
[1] 98[1] 111Mismatched Data Sets with Common Keys
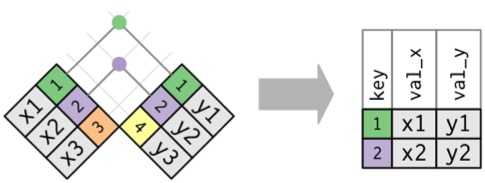
Mismatched Data Sets with Common Keys
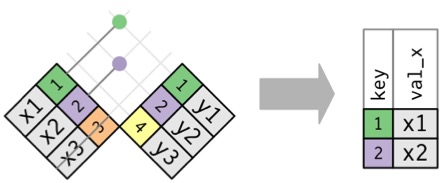
Inner Join
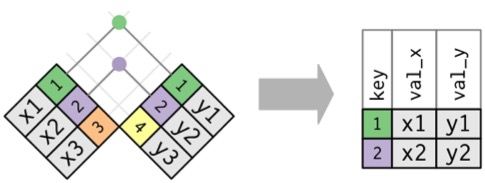
Creates new Data with rows that exist in both data sets
Reducing Data in Inner Joins
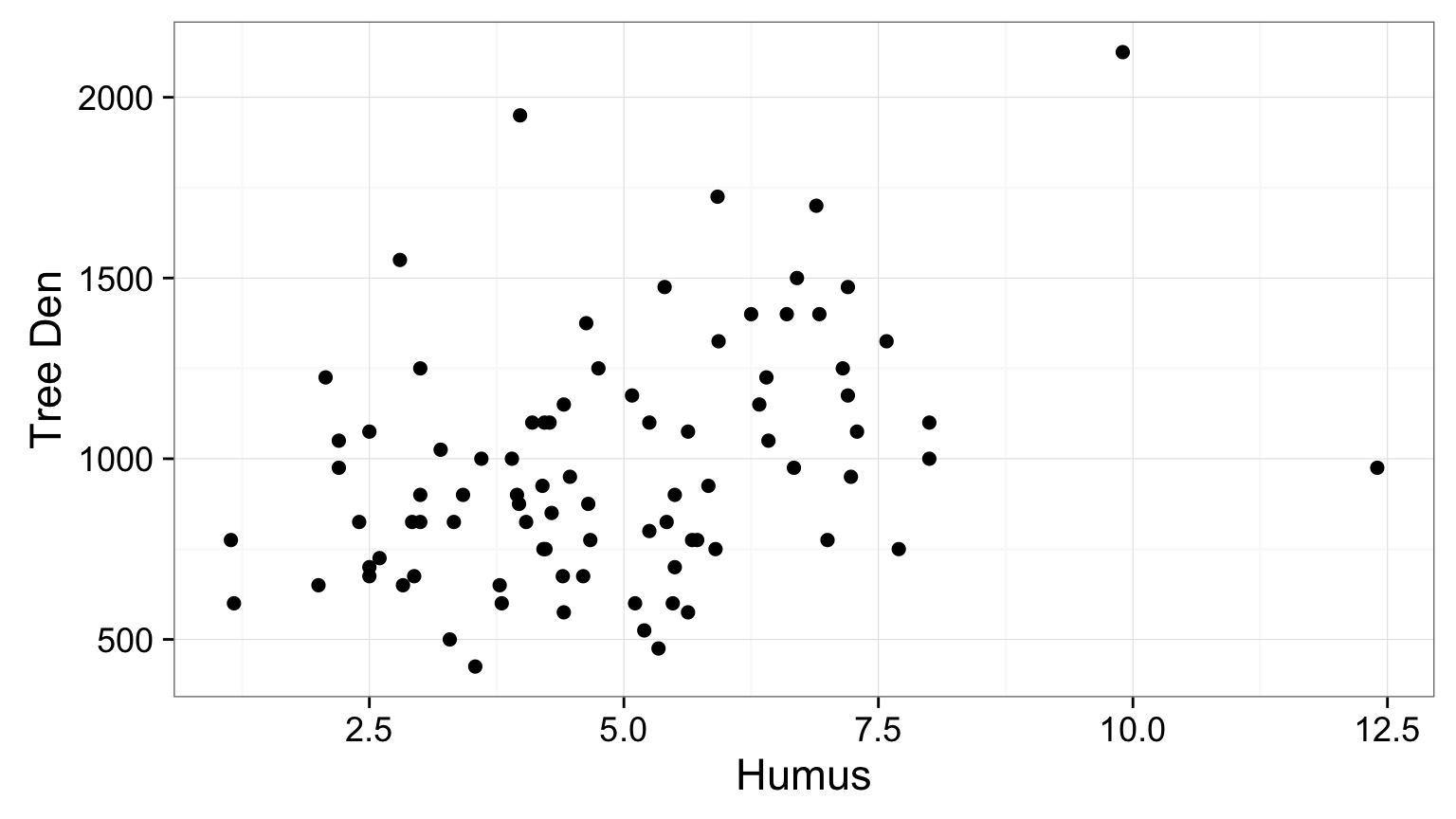
Joining with `by = join_by(Stand, Year, Longitude, Latitude)`Plotting Paired Data
Outer Joins
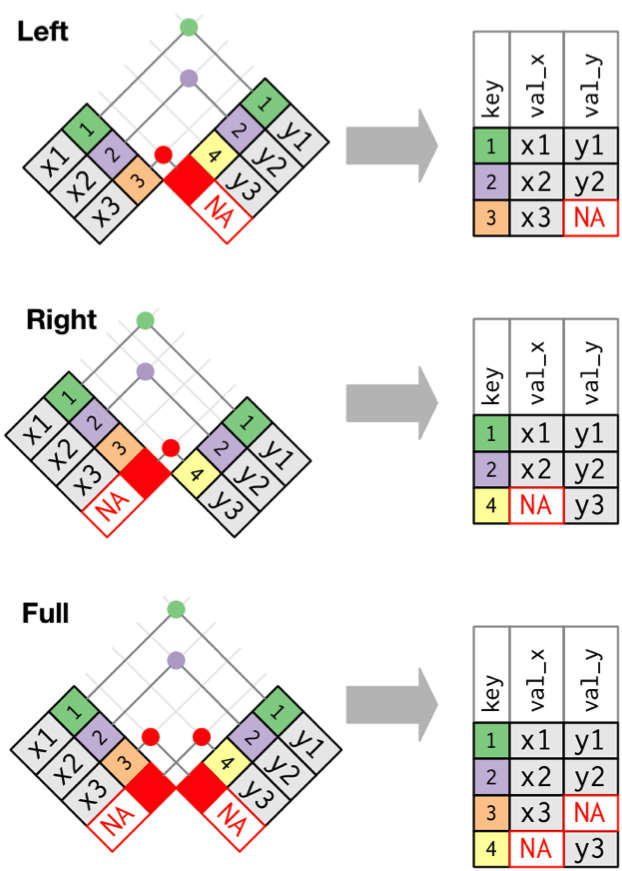
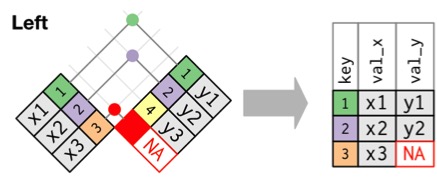
Left Join: Retain Rows with NAs in First Dataset
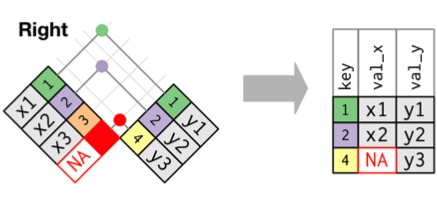
Joining with `by = join_by(Stand, Year, Longitude, Latitude)`Right Join: Retain Rows with NAs in Second Dataset
Joining with `by = join_by(Stand, Year, Longitude, Latitude)`Full Join: Bring it All Together
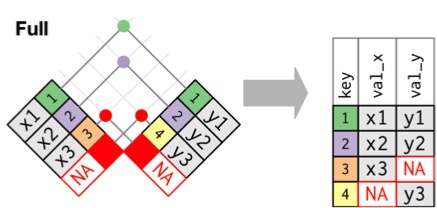
Good to see the full dataset
Joining with `by = join_by(Stand, Year, Longitude, Latitude)`The Joins
Filtering Joins
- I only want data that matches a set of criteria
- Like outer joins with a second na.omit step
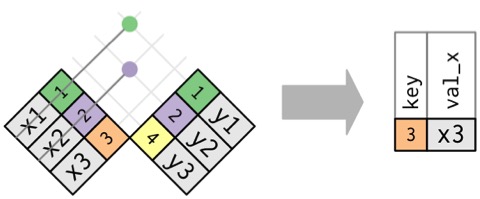
Semi Join: X %in% Y
Good before data pre-processing
Joining with `by = join_by(Stand, Year, Longitude, Latitude)`Anti Join: X NOT %in% Y
Good for diagnosing data mismatch
Joining with `by = join_by(Stand, Year, Longitude, Latitude)`Exercise 1
- You want to plot a map of the sites
- You want size of points to be area
- You want color of points to be dead Hemlock area
Exercise 2
- You want to plot a map of the sites
- BUT - you want to show which sites are missing environmental
data
- AND - you want to show which sites are missing biological data
- (this might be more than one plot and more than one data join!)