class: middle, center background-color: #a95c68 # Sampling Estimates, Precision, and Simulation  ### Biol 607 --- # Today's Etherpad <br><br><br>.center[.large[https://etherpad.wikimedia.org/p/sampling-2020]] --- # Estimation and Precision .large[ 1. Probability Distributions and Population Parameter Estimates 2. Simulation, Precision, and Sample Size Estimation 3. Bootstrapping our Way to Confidence ] --- # Last Time: Sample Versus Population <img src="04_simulation_estimation_x_files/figure-html/samp_pop_plot-1.png" style="display: block; margin: auto;" /> --- # Sample Properties: **Mean** `$$\bar{x} = \frac{ \displaystyle \sum_{i=1}^{n}{x_{i}} }{n}$$` `\(\large \bar{x}\)` - The average value of a sample `\(x_{i}\)` - The value of a measurement for a single individual n - The number of individuals in a sample `\(\mu\)` - The average value of a population (Greek = population, Latin = Sample) --- class: center, middle # Our goal is to get samples representative of a population, and estimate population parameters. We assume a **distribution** of values to the population. --- # Probability Distributions <img src="04_simulation_estimation_x_files/figure-html/normplot-1.png" style="display: block; margin: auto;" /> --- # Probability Distributions Come in Many Shapes <img src="04_simulation_estimation_x_files/figure-html/dists-1.png" style="display: block; margin: auto;" /> --- # The Normal (Gaussian) Distribution <img src="04_simulation_estimation_x_files/figure-html/normplot-1.png" style="display: block; margin: auto;" /> - Arises from some deterministic value and many small additive deviations - VERY common --- class: center # Understanding Gaussian Distributions with a Galton Board (Quinqunx) <video controls loop><source src="04_simulation_estimation_x_files/figure-html/quincunx.webm" /></video> --- # We see this pattern everywhere - the Random or Drunkard's Walk ```r one_path <- function(steps){ each_step <- c(0, runif(steps, min = -1, max = 1)) path <- cumsum(each_step) return(path) } ``` -- 1. Input some number of steps to take 2. Make a vector of 0 and a bunch of random numbers from -1 to 1 3. Take the cummulative sum across the vector to represent the path 4. Return the path vector --- # 1000 Simulated Random Walks to Normal Homes <img src="04_simulation_estimation_x_files/figure-html/all_walks-1.png" style="display: block; margin: auto;" /> --- # A Normal Result for Final Position <img src="04_simulation_estimation_x_files/figure-html/final_walk-1.png" style="display: block; margin: auto;" /> --- # Normal distributions <img src="04_simulation_estimation_x_files/figure-html/normplot-1.png" style="display: block; margin: auto;" /> - Results from additive accumulation of many small errors - Defined by a mean and standard deviation: `\(N(\mu, \sigma)\)` - 2/3 of data is within 1 SD of the mean - Values are peaked without **skew** (skewness = 0) - Tails are neither too thin nor too fat (**kurtosis** = 0) --- # Estimation and Precision .large[ 1. Probability Distributions and Population Parameter Estimates 2. .red[Simulation, Precision, and Sample Size Estimation] 3. Bootstrapping our Way to Confidence ] --- class: middle, center # The Eternal Question: What should my sample size be? --- # Let's find out .large[ 1. Get in groups of 3 <br><br> 2. Ask each other your age. Report the mean to me.<br><br> 3. Now get into another group of five, and do the same.<br><br> 4. Now get into another group of ten, and do the same.<br><br> ] --- class: center, middle # We simulated sampling from our class population! --- # What if We Could Pretend to Sample? .large[ - Assume the distribution of a population - Draw simulated 'samples' from the population at different sample sizes - Examine when an estimated property levels off or precision is sufficient - Here we define Precision as 1/variance at a sample size ] --- background-image: url(images/04/is-this-a-simulation.jpg) background-position: center background-size: cover --- background-image: url(images/04/firefly-ship.jpg) background-position: center background-size: cover class: center # .inverse[Let's talk Firefly] --- background-image: url(images/04/fireflies-1500x1000.jpg) background-position: center background-size: cover --- # Start With a Population... Mean of Firefly flashing times: 95.9428571 SD of Firefly flasing times: 10.9944982 -- So assuming a normal distribution... -- <img src="04_simulation_estimation_x_files/figure-html/fireflydist-1.png" style="display: block; margin: auto;" /> --- # Choose a Random Sample - n=5? Mean of Firefly flashing times: 95.9428571 SD of Firefly flasing times: 10.9944982 So assuming a normal distribution... <img src="04_simulation_estimation_x_files/figure-html/fireflydistPoints-1.png" style="display: block; margin: auto;" /> -- ```r rnorm(n = 5, mean = mean(firefly$flash.ms), sd = sd(firefly$flash.ms) ``` --- # Calculate Sample Mean Mean of Firefly flashing times: 95.9428571 SD of Firefly flasing times: 10.9944982 So assuming a normal distribution... <img src="04_simulation_estimation_x_files/figure-html/fireflydistMean-1.png" style="display: block; margin: auto;" /> -- Rinse and repeat... --- # How Good is our Sample Size for Estimating a Mean? <img src="04_simulation_estimation_x_files/figure-html/plot_dist_sim-1.png" style="display: block; margin: auto;" /> --- # Where does the variability level off? <img src="04_simulation_estimation_x_files/figure-html/dist_sim_stop-1.png" style="display: block; margin: auto;" /> --- # Many Ways to Visualize <img src="04_simulation_estimation_x_files/figure-html/bin2d-1.png" style="display: block; margin: auto;" /> --- # Where does the variability level off? ``` # A tibble: 99 x 2 sampSize mean_sim_sd <int> <dbl> 1 96 1.10 2 100 1.11 3 92 1.12 4 95 1.12 5 97 1.13 6 89 1.13 7 93 1.13 8 98 1.13 9 99 1.14 10 91 1.17 # … with 89 more rows ``` --- # Visualize Variability in Estimate of Mean <img src="04_simulation_estimation_x_files/figure-html/sim_prec-1.png" style="display: block; margin: auto;" /> -- .large[What is acceptable to you? And/or relative to the Mean?] --- class: center, middle # **Central Limit Theorem** The distribution of means of a sufficiently large sample size will be approximately normal https://istats.shinyapps.io/sampdist_cont/ --- class: center, middle # The Standard Error <br><br> .large[ A standard error is the standard deviation of an estimated parameter if we were able to sample it repeatedly. ] --- # But, I only Have One Sample? How can I know my SE for my mean or any other parameter? <img src="04_simulation_estimation_x_files/figure-html/firehist-1.png" style="display: block; margin: auto;" /> --- # Estimation and Precision .large[ 1. Probability Distributions and Population Parameter Estimates 2. Simulation, Precision, and Sample Size Estimation 3. .red[Bootstrapping our Way to Confidence] ] --- # The Bootstrap .large[ - We can resample our sample some number of times with replacement - This resampling with replacement is called **bootstrapping** - One replicate simulation is one **bootstrap** ] --- # One Bootstrap Sample in R ```r # One bootstrap sample(firefly$flash.ms, size = nrow(firefly), replace = TRUE) ``` ``` [1] 95 95 85 87 86 95 80 109 86 95 106 96 109 88 86 87 112 92 79 [20] 116 89 92 98 112 113 118 94 94 95 86 87 94 118 109 98 ``` --- # Boostrapped Estimate of a SE - We can calculate the Standard Deviation of a sample statistic from replicate bootstraped samples - This is called the botstrapped **Standard Error** of the estimate ```r one_boot <- function(){ sample(firefly$flash.ms, size = nrow(firefly), replace = TRUE) } boot_means <- replicate(1000, mean(one_boot())) sd(boot_means) ``` ``` [1] 1.816216 ``` --- class: large # So I always have to boostrap Standard Errors? -- ## .center[**No**] -- Many common estimates have formulae, e.g.: `$$SE_{mean} = \frac{s}{\sqrt(n)}$$` -- Boot SEM: Boot SEM: 1.816, Est. SEM: 2 -- .center[(but for medians, etc., yes )] --- # Bootstrapping to Estimate Precision of a Non-Standard Metric <img src="04_simulation_estimation_x_files/figure-html/unnamed-chunk-4-1.png" style="display: block; margin: auto;" /> SE of the SD = 1.101 --- class: large # Standard Error as a Measure of Confidence -- .center[.red[**Warning: this gets weird**]] -- - We have drawn our SE from a **sample** - not the **population** -- - Our estimate ± 1 SE tells us 2/3 of the **estimates** we could get by resampling this sample -- - This is **not** 2/3 of the possible **true parameter values** -- - Rather, if we were to sample the population many many times, 2/3 of the time, this interval will contain the true value --- # Confidence Intervals .large[ - So, 1 SE = the 66% Confidence Interval - ~2 SE = 95% Confidence Interval - Best to view these as a measure of precision of your estimate - And remember, if you were able to do the sampling again and again and again, some fraction of your intervals would contain a true value ] --- class: large, middle # Let's see this in action ### .center[.middle[https://istats.shinyapps.io/ExploreCoverage/]] --- # Frequentist Philosophy .large[The ideal of drawing conclusions from data based on properties derived from theoretical resampling is fundamentally **frequentist** - i.e., assumes that we can derive truth by observing a result with some frequency in the long run.] <img src="04_simulation_estimation_x_files/figure-html/CI_sim-1.png" style="display: block; margin: auto;" /> --- # A Final Note: SE, SD, CIs.... 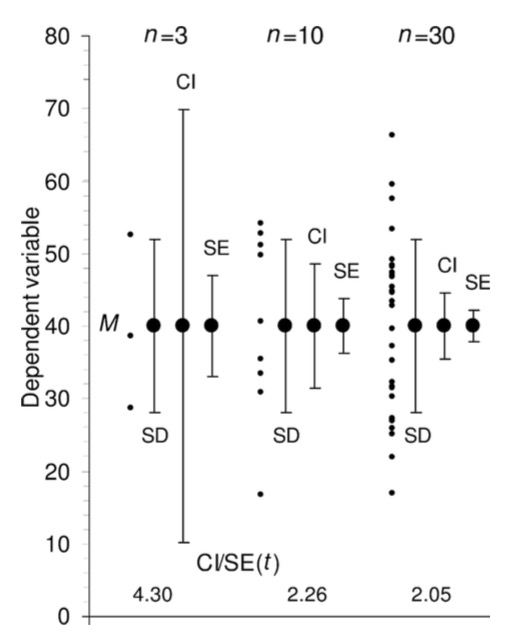 .bottom[.small[.left[[Cumming et al. 2007 Figure 4](http://byrneslab.net/classes/biol-607/readings/Cumming_2007_error.pdf)]]] --- # SE, SD, CIs.... 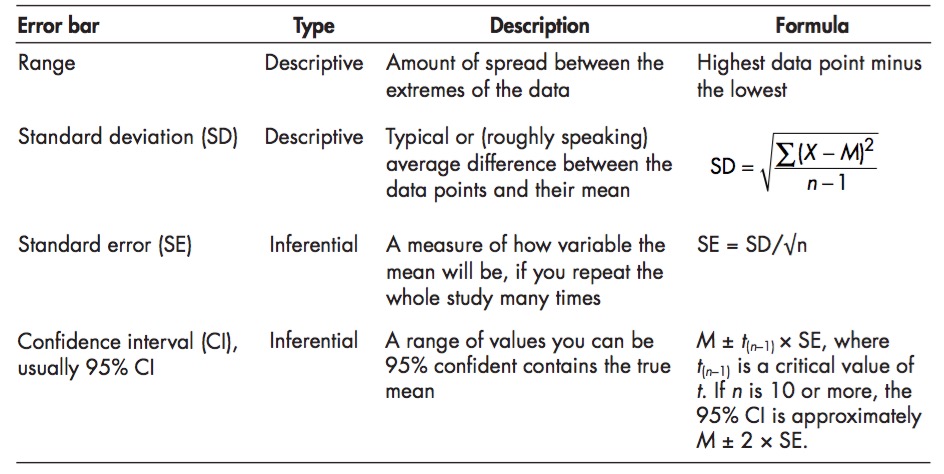 .bottom[.small[.left[[Cumming et al. 2007 Table 1](http://byrneslab.net/classes/biol-607/readings/Cumming_2007_error.pdf)]]]